On July 2, Nature Communications, an international authoritative journal, published online the latest research results of Professor Wang Jianxin 's team from the School of Computer Science and Engineering, Central South University (CSU), “HiTE: a fast and accurate dynamic boundary adjustment approach for full-length transposable elements detection and annotation”. In this paper, a new method for transposon detection and annotation based on genome assembly data was proposed, and the corresponding software HiTE was developed. Hu Kang and Ni Peng from School of Computer Science and Engineering of CSU were the co-first authors of the paper, Professor Wang Jianxin from School of Computer Science and Engineering of CSU was one of the co-corresponding authors of the paper, and Central South University was the first authorship institute. This study was supported by many projects such as the National Key Research and Development Program of China, the National Natural Science Foundation of China, and the Xiangjiang Laboratory Projects of Opening Competition Mechanism to Select the Best Candidates.
Transposable elements (TEs) make up the majority of repetitive regions in most eukaryotic species and are known to have a significant impact on genome evolution and intraspecific genomic diversity. TEs have been found to play a key role in human disease and crop breeding by interrupting or regulating key genes. In recent years, advancements in genome assembly technology have greatly improved the prospects for comprehensive annotation of transposable elements. However, existing methods for TE annotation using genome assemblies suffer from limited accuracy and robustness, requiring extensive manual edition. In addition, the currently available gold-standard TE databases are not comprehensive, even for extensively studied species, highlighting the critical need for an automated TE detection method to supplement existing repositories.
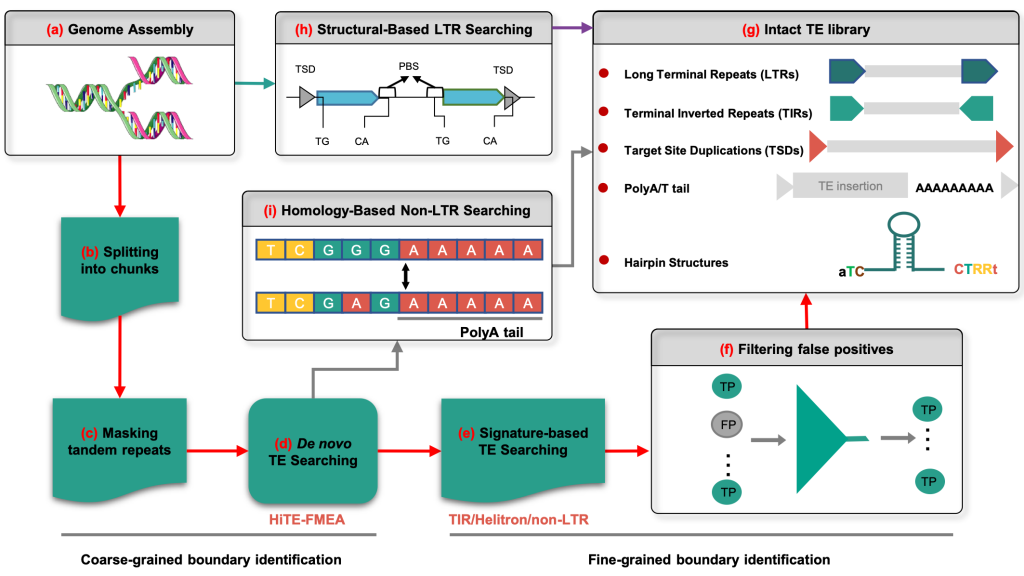
Workflow of the HiTE pipeline for TE Annotation
In view of this limitation, Professor Wang Jianxin's team introduced a fast and accurate dynamic boundary adjustment approach designed to detect full-length TEs by deeply analyzing the sequence structure characteristics and biological repetitive nature of transposable elements. The number of full-length TEs identified by HiTE was far more than that identified by the existing tools using a common 95% assessment threshold, such as 1078 full-length TE sequences identified in rice using HiTE, compared with 506 and 378 identified by state-of-the-art tools EDTA and RepeatModeler2, respectively. In order to solve the problem of high false-positive in transposon identification, a variety of highly reliable filtering algorithms were implemented, which can filter the majority of false-positive sequences. The experimental results showed that the filtered HiTE obtained a higher number of full-length TEs with significant improvement of the accuracy in TEs identification without reducing the sensitivity, and achieved F1 scores of 93.56%, which were much higher than 87.34% and 54.82% attained by EDTA and RepeatModeler2.
(First Reviewer: Dai Yu'ou, Second Reviewer: Deng Haodi, Third Reviewer: Li Yin)
Source: School of Computer Science and Engineering, Author: Hu Kang
Original article link: https://news.csu.edu.cn/info/1003/159034.htm











